ontology
A rough translation of terms used to describe relational databases and ontologies:
| Relational database | Ontology |
| row | subject |
| column | predicate |
| table data | literal nodes |
OWL Synopsis
- minCardinality (only 0 or 1)
- maxCardinality (only 0 or 1)
- cardinality (only 0 or 1)
SPARQL
SPARQL is a query language for ontologies
SELECT ?name
WHERE
{
[ a my:person;
my:suffers my:insomnia;
my:name ?name].
}
This query returns the names of all people who suffer insomnia. It uses a syntax similar to SQL and contains a blank node inside a WHERE clause.
SPARQL supports keywords like (NOT) EXISTS, UNION, OPTIONAL, DISTINCT, MINUS, GROUP BY, HAVING. Available aggregates are: COUNT, SUM, MIN, MAX, AVG, GROUP_CONCAT, and SAMPLE. Moreover, all XPath functions (e.g., string manipulation utils like concat) can be used as long as they are preceded with the "xsd:" prefix.
SELECT (SUM(?lprice) AS ?totalPrice)
WHERE {
?org :affiliates ?auth .
?auth :writesBook ?book .
?book :price ?lprice .
}
GROUP BY ?org
HAVING (SUM(?lprice) > 10)
DELETE
{ ?x my:likes ?y . }
WHERE
{ ?x a my:person .
?y a my:person; my:suffers my:xenophobia .
}
Removes people that like xenophobia
INSERT
{ ?x my:loves ?y . }
WHERE
{
?x a my:person .
MINUS {?x a my:girl}
?y a my:person, my:girl .
}
Outputs that everybody loves girls -- excluding the girls themselves
dotNetRDF Sample
N3 Notation:
@prefix my: <http://www.codeproject.com/KB/recipes/n3_notation#>.
my:Peter a my:person, my:boy;
my:suffers my:acrophobia, my:insomnia, my:xenophobia;
my:name "Peter";
my:likes my:Kate.
my:Mark a my:person, my:boy;
my:suffers my:insomnia;
my:name "Mark".
my:Kate a my:person, my:girl;
my:name "Kate".
SparQL statement:
PREFIX my: <http://www.codeproject.com/KB/recipes/n3_notation#>
SELECT ?name
WHERE {
[ a my:person;
my:suffers my:insomnia;
my:name ?name] .
}
SparQL Results:
- Peter
- Mark
ChEBI
Chemical Entities of Biological Interest, also known as ChEBI, is a database and ontology of molecular entities focused on 'small' chemical compounds, that is part of the Open Biomedical Ontologies effort.
ChEBI ontology files ChEBI schema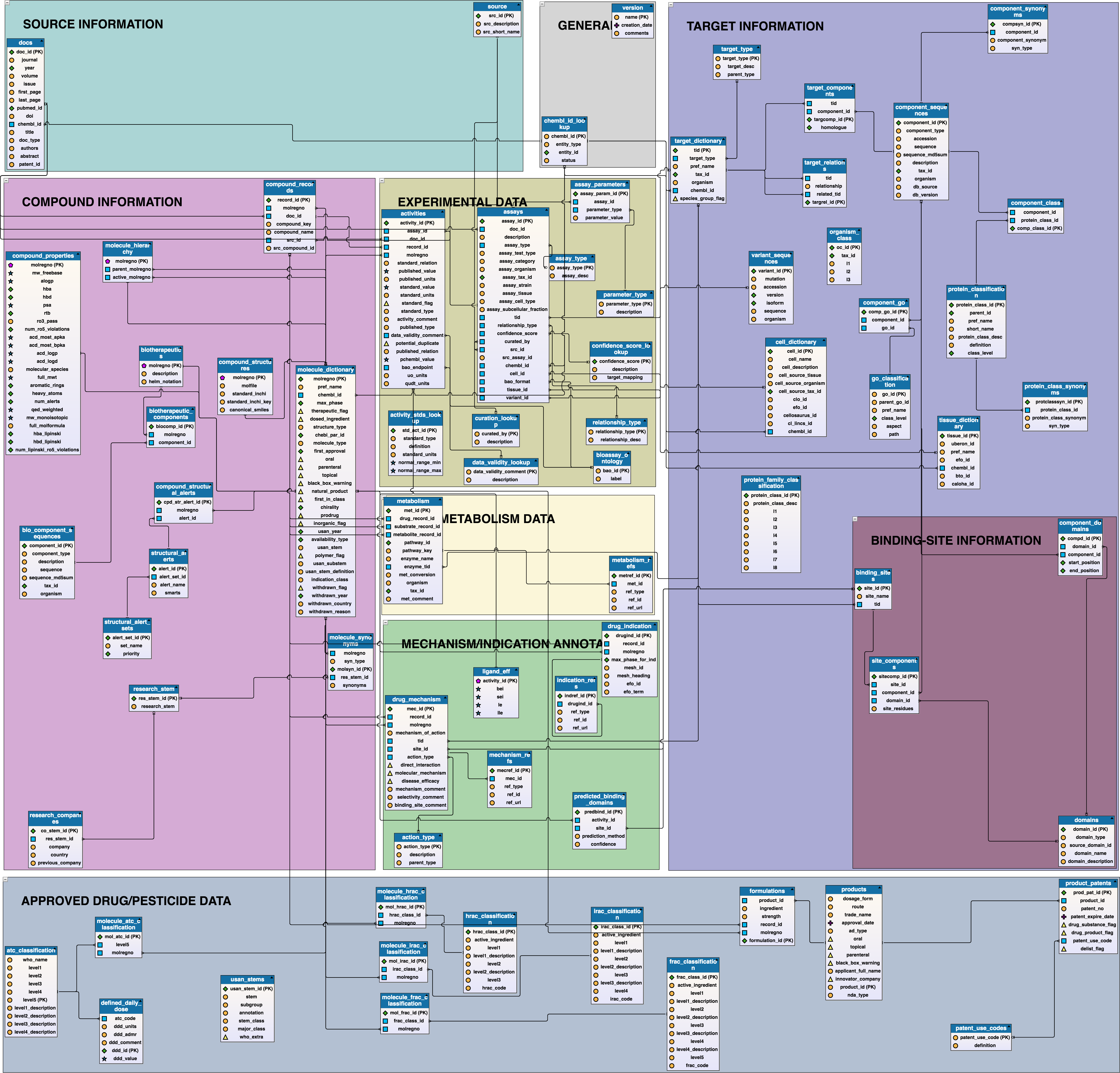
FoodOn
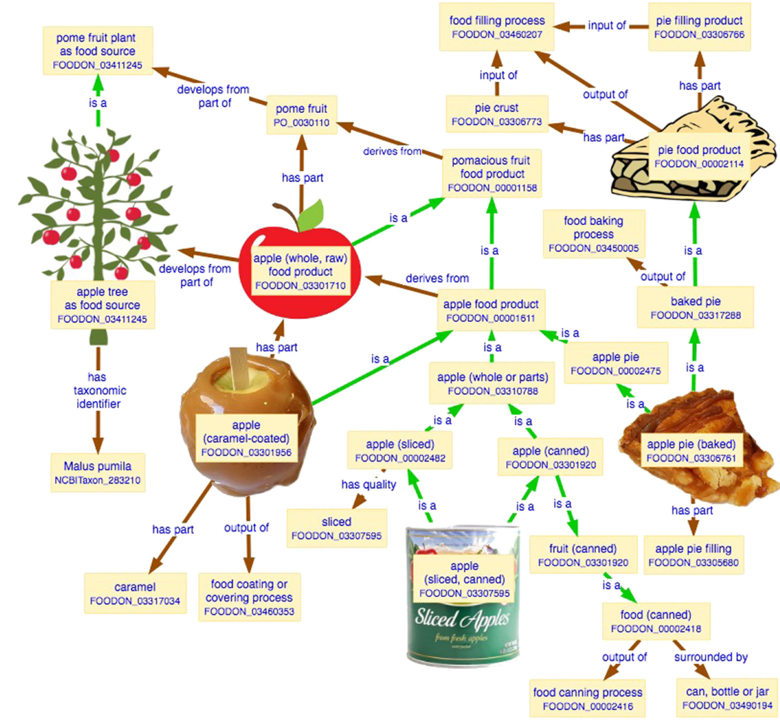
2018-09-09
FOODON
FoodOn (http://foodon.org) is a consortium-driven project to build a comprehensive and easily accessible global farm-to-fork ontology about food, that accurately and consistently describes foods commonly known in cultures from around the world.
Food Ontology
GTIN
EAN全名为European Article Number(欧洲商品条码),在1977年时由欧洲几个主要工业国家共同发展出来的,后来变成国际商品条码系统。台湾在1985年加入EAN会员,现在我们买东西时在柜台结帐,服务人员用扫瞄器所读的商品上的条码就是EAN条码。国际商品条码需经过申请,不可自行编码列印。
EAN-13现在称之为GTIN-13 (Global Trade Item Number)条码,属于GS1国际条码系统其中的一种。
备注:GS1国际条码系统包括GTIN-13、GTIN-12(原称UPC)、GTIN-8(原称EAN-8)、GLN、GTIN-14(ITF-14)、SSCC、GS1-128(原称EAN- 128)、GS1 Databar(原称RSS code)、GS1 Data Matrix 等。
KEGG MEDICUS
KEGG MEDICUS is an integrated information resource of diseases, drugs, and health-related substances, aiming to bring the genomic revolution to society (see background information). Specifically, package insert information for all drugs marketed in Japan and the USA is integrated with KEGG DRUG and KEGG DISEASE.
The KEGG MEDICUS subset is freely available at the GenomeNet FTP site.
ftp://ftp.genome.jp/pub/kegg/medicus/
plant environment ontology
plant environment ontology (EO:0007359): A set of standardized controlled vocabularies to describe various types of treatments given to an individual plant / a population or a cultured tissue and/or cell type sample to evaluate the response on its exposure. It also includes the study types, where the terms can be used to identify the growth study facility. Each growth facility such as field study, growth chamber, green house etc is a environment on its own it may also involve instances of biotic and abiotic environments as supplemental treatments used in these studies.
plant environment (EO:0007359): A plant treatment, growing condition, and/or study type applied to a whole plant (PO:0000003), a plant structure (PO:00090119), and/or a plant population to evaluate the plant response.
Revised comment: The Plant Environment Ontology describes the treatments, growing conditions, and/or study types used in various types of plant biology experiments. The subjects of the studies may also include in vitro plant structures (PO:0000004). A plant treatment is the most specific, and may be a component of a plant growing condition. The broadest classes are the plant study types where the terms can be used to identify the growth study facility. Each growth facility such as field study, growth chamber, green house etc is a environment on its own it may also involve instances of biotic and abiotic treatments and different growing conditions.
Taxonomy vs Ontology
taxonomy is usually only a hierarchy of concepts (i.e. the only relation between the concepts is parent/child, or subClass/superClass, or broader/narrower)
in an ontology, arbitrary complex relations between concepts can be expressed too (X marriedTo Y; or A worksFor B; or C locatedIn D, etc )
Knowledge Graph 知识图谱
An Ontology captures the entities, their types(classes), class hierarchy,properties and relationships between the entities. An Ontology has instances/individuals. But, if you are dealing with data , then a Knowledge Graph is a better option. The data/triples are stored in the form of nodes and edges,i.e; each data point is a node and the relationship becomes an edge that connects those nodes.
So, you can say an Ontology records the structure/schema whereas a Knowledge Graph captures the data. Moreover, an ontology has the power to enable restrictions/rules and reasoning.
Assume "a knowledge graph" is a Semantic Network. Google Knowledge Graph is a Semantic Network.
An OWL Ontology provides classification - kind-of dynamic schema - for a Semantic Network. So, in your terms, an ontology would be metadata for a knowledge graph.
OBO Stanza Conventions
Header Tags
Header tags should appear in the following order:
format-version
data-version
date
saved-by
auto-generated-by
import
subsetdef
synonymtypedef
default-namespace
namespace-id-rule
idspace
treat-xrefs-as-equivalent
treat-xrefs-as-genus-differentia
treat-xrefs-as-relationship
treat-xrefs-as-is_a
remark
ontology
Ordering Term and Typedef stanzas
[Term], [Typdef], and [Instance] stanzas should be serialized in alphabetical order on the value of their id tag.
Ordering Term and Typedef tags
Term tags should appear in the following order:
id
is_anonymous
name
namespace
alt_id
def
comment
subset
synonym
xref
builtin
property_value
is_a
intersection_of
union_of
equivalent_to
disjoint_from
relationship
created_by
creation_date
is_obsolete
replaced_by
consider
Typedef tags should appear in the following order:
id
is_anonymous
name
namespace
alt_id
def
comment
subset
synonym
xref
property_value
domain
range
builtin
holds_over_chain
is_anti_symmetric
is_cyclic
is_reflexive
is_symmetric
is_transitive
is_functional
is_inverse_functional
is_a
intersection_of
union_of
equivalent_to
disjoint_from
inverse_of
transitive_over
equivalent_to_chain
disjoint_over
relationship
is_obsolete
created_by
creation_date
replaced_by
consider
expand_assertion_to
expand_expression_to
is_metadata_tag
is_class_level
Instance tags should appear in the following order:
id
is_anonymous
name
namespace
alt_id
def
comment
subset
synonym
xref
instance_of
property_value
relationship
created_by
creation_date
is_obsolete
replaced_by
consider
Reference: https://owlcollab.github.io/oboformat/doc/GO.format.obo-1_4.html
常见Metadata标准
网络资源:Dublin Core、IAFA Template、CDF、Web Collections
文献资料:MARC(with 856 Field),Dublic Core
人文科学:TEI Header
社会科学数据集:ICPSR SGML Codebook
博物馆与艺术作品:CIMI、CDWA、RLG REACH Element Set、VRA Core
政府信息:GILS
地理空间信息:FGDC/CSDGM
数字图像:MOA2 metadata、CDL metadata、Open Archives Format、VRA Core、NISO/CLIR/RLG Technical Metadata for Images
档案库与资源集合:EAD
技术报告:RFC 1807
连续图像:MPEG-7
ProteoWizard msconvert supported formats
| Vendor | Formats | Vendor Required Software |
|---|---|---|
| ABI | T2D | DataExplorer 4.0 |
| Agilent | MassHunter .d | distributed with ProteoWizard |
| Bruker | Compass .d, YEP, BAF, FID, TDF | distributed with ProteoWizard |
| Sciex | WIFF / WIFF2 | distributed with ProteoWizard |
| Shimadzu | LCD | distributed with ProteoWizard |
| Thermo Scientific | RAW | distributed with ProteoWizard |
| Waters | MassLynx .raw / UNIFI | distributed with ProteoWizard |
Attribute & Property
These words existed way before Computer Science came around.
Attribute is a quality or object that we attribute to someone or something. For example, the scepter is an attribute of power and statehood.
Property is a quality that exists without any attribution. For example, clay has adhesive qualities; or, one of the properties of metals is electrical conductivity. Properties demonstrate themselves though physical phenomena without the need attribute them to someone or something. By the same token, saying that someone has masculine attributes is self-evident. In effect, you could say that a property is owned by someone or something.
To be fair though, in Computer Science these two words, at least for the most part, can be used interchangeably - but then again programmers usually don't hold degrees in English Literature and do not write or care much about grammar books :).
mzML file format specification
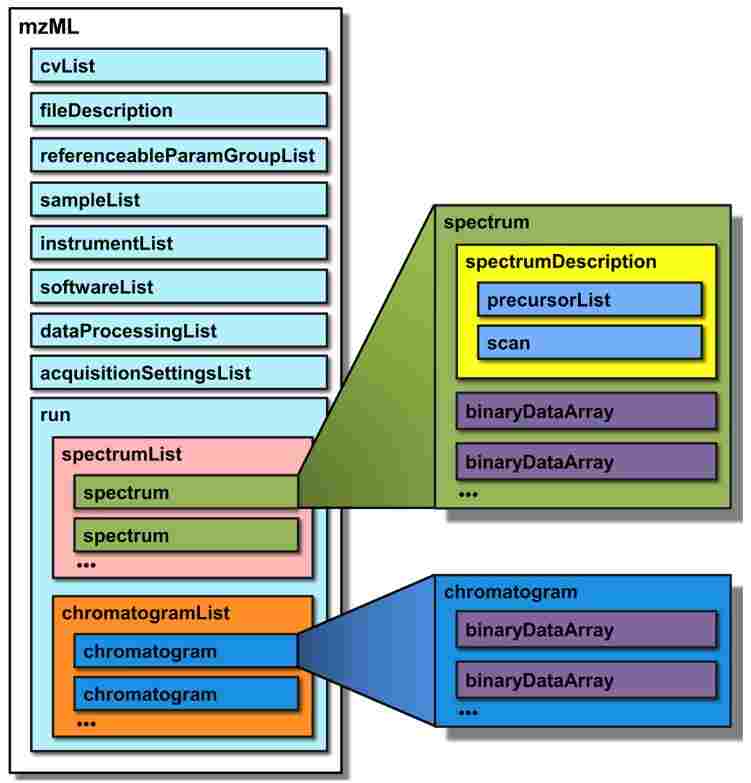
cvList contains information about the controlled vocabularies referenced in the rest of the mzML document; fileDescription contains basic information on the type of spectra contained in the file; referenceableParamGroupList is an optional element that of groups of controlled vocabulary terms that can be referenced as a unit throughout defines a list the document; sampleList can contain information about samples that are referenced in the file; instrumentConfigurationList contains information about the instrument that generated the run; softwareList and dataProcessingList provide a history of data processing that occurred after the raw acquisition; acquisitionSettingsList is an optional element that stores special input parameters for the mass spectrometer, such as inclusion lists. These elements are followed by the acquired spectra and chromatograms. Both spectral and chromatographic data are represented by binary format data encoded into base 64 strings, rather than human-readable ASCII text for enhanced fidelity and efficiency when dealing with profile data. This design choice does not enjoy unanimous approval, but has been agreed upon by the majority of designers
Martens L, Chambers M C, Sturm M, et al. mzML—a Community Standard for Mass Spectrometry Data [J]. Molecular & Cellular Proteomics, 2011, 10(1): 1-7