| E0065 |
SPACS |
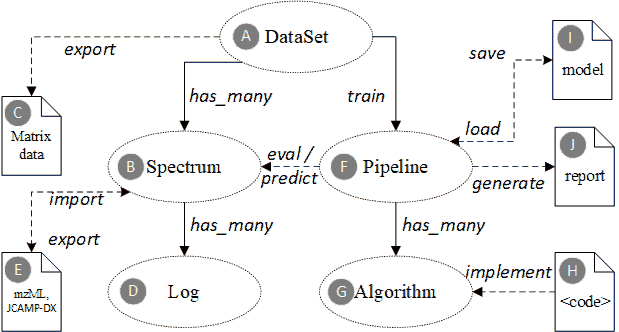
DataSet |
[Entity] |
34E5E14 |
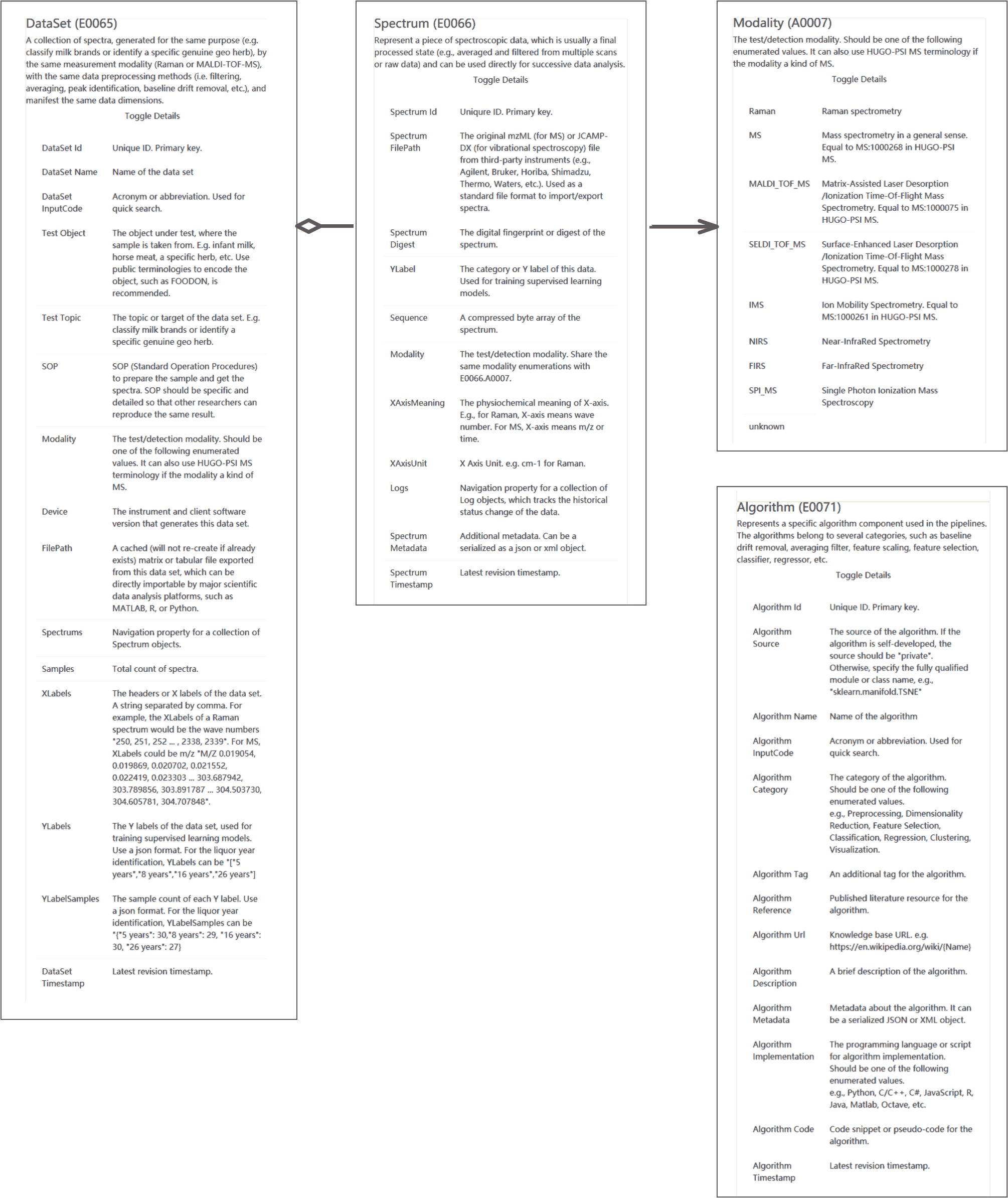
A collection of spectrum data, generated for the same purpose (e.g. classify milk brands or identify a specific genuine geo herb), by the same measurement modality (Raman or MALDI-TOF-MS), with the same data preprocessing methods (i.e. filtering, averaging, peak identification, baseline drift removal, etc.), and manifest the same data dimensions. |
| E0065.A0001 |
SPACS |
DataSet Id |
[Attribute] |
1F6323 |
Unique ID. Primary key. |
| E0065.A0002 |
SPACS |
DataSet Name |
[Attribute] |
7DA012 |
Name of the data set |
| E0065.A0003 |
SPACS |
DataSet InputCode |
[Attribute] |
20AB2F8 |
Acronym or abbreviation. Used for quick search. |
| E0065.A0004 |
SPACS |
Test Object |
[Attribute] |
11343B8 |
The object under test, where the sample is taken from. E.g. infant milk, horse meat, a specific herb, etc. Use public terminologies to encode the object, such as FOODON, is recommended. |
| E0065.A0005 |
SPACS |
Test Topic |
[Attribute] |
F11A3A |
The topic or target of the data set. E.g. classify milk brands or identify a specific genuine geo herb. |
| E0065.A0006 |
SPACS |
SOP |
[Attribute] |
19426AF |
SOP (Standard Operation Procedures) to prepare the sample and get the spectrum data. SOP should be specific and detailed so that other researchers can reproduce the same result. |
| E0065.A0007 |
SPACS |
Modality |
[Attribute] |
25C0BD0 |
The test/detection modality. Should be one of the following enumerated values. Can also use HUGO-PSI MS terminology if the modality a kind of MS. |
| E0065.A0007.V0001 |
SPACS |
Raman |
[Value] |
1483B1F |
Raman spectrometry |
| E0065.A0007.V0002 |
SPACS |
MS |
[Value] |
3C53323 |
Mass spectrometry in a general sense. Equal to MS:1000268 in HUGO-PSI MS. |
| E0065.A0007.V0003 |
SPACS |
MALDI_TOF_MS |
[Value] |
321EFAE |
Matrix-Assisted Laser Desorption /Ionization Time-Of-Flight Mass Spectrometry. Equal to MS:1000075 in HUGO-PSI MS. |
| E0065.A0007.V0004 |
SPACS |
SELDI_TOF_MS |
[Value] |
2211C1A |
Surface-Enhanced Laser Desorption /Ionization Time-Of-Flight Mass Spectrometry. Equal to MS:1000278 in HUGO-PSI MS. |
| E0065.A0007.V0005 |
SPACS |
IMS |
[Value] |
461787 |
Ion Mobility Spectrometry. Equal to MS:1000261 in HUGO-PSI MS. |
| E0065.A0007.V0006 |
SPACS |
NIRS |
[Value] |
8E5B1E |
Near-InfraRed Spectrometry |
| E0065.A0007.V0007 |
SPACS |
FIRS |
[Value] |
35C5250 |
Far-InfraRed Spectrometry |
| E0065.A0007.V0008 |
SPACS |
SPI_MS |
[Value] |
2913516 |
Single Photon Ionization Mass Spectroscopy |
| E0065.A0007.V0009 |
SPACS |
unknown |
[Value] |
4FF431 |
|
| E0065.A0008 |
SPACS |
Device |
[Attribute] |
2C3C76B |
The instrument and client software version that generates this data set. |
| E0065.A0009 |
SPACS |
FilePath |
[Attribute] |
A96DD3 |
A cached (will not re-create if already exists) matrix or tabular file exported from this data set, which can be directly importable by major scientific data analysis platforms, such as MATLAB, R, or Python. |
| E0065.A0010 |
SPACS |
Spectrums |
[Attribute] |
362B004 |
Navigation property for a collection of Spectrum objects. |
| E0065.A0011 |
SPACS |
Samples |
[Attribute] |
128345D |
Total count of spectrum data samples. |
| E0065.A0012 |
SPACS |
XLabels |
[Attribute] |
29E3549 |
The headers or X labels of the data set. A string separated by comma. For example, the XLabels of a Raman spectrum data would be the wave numbers "250, 251, 252 ... , 2338, 2339". For MS, XLabels could be m/z "M/Z 0.019054, 0.019869, 0.020702, 0.021552, 0.022419, 0.023303 ... 303.687942, 303.789856, 303.891787 ... 304.503730, 304.605781, 304.707848". |
| E0065.A0013 |
SPACS |
YLabels |
[Attribute] |
1C52978 |
The Y labels of the data set, used for training supervised learning models. Use a json format. For the liquor year identification, YLabels can be "["5 years","8 years","16 years","26 years"] |
| E0065.A0014 |
SPACS |
YLabelSamples |
[Attribute] |
117EE24 |
The sample count of each Y label. Use a json format. For the liquor year identification, YLabelSamples can be "{"5 years": 30,"8 years": 29, "16 years": 30, "26 years": 27} |
| E0065.A0015 |
SPACS |
DataSet Timestamp |
[Attribute] |
1C5834C |
Latest revision timestamp. |
| E0066 |
SPACS |
Spectrum |
[Entity] |
174E9FA |
Represent a piece of spectroscopic data, which is usually a final processed state (e.g. averaged and filtered from multiple scans or raw data) and can be used directly for successive data analysis. |
| E0066.A0001 |
SPACS |
Spectrum Id |
[Attribute] |
2063480 |
Uniqure ID. Primary key. |
| E0066.A0002 |
SPACS |
Spectrum FilePath |
[Attribute] |
6C58B5 |
The original mzML (for MS) or JCAMP-DX (for vibrational spectroscopy) file from third-party instruments (e.g. Agilent, Bruker, Horiba, Shimadzu, Thermo, Waters, etc.). Used as a standard file format to import/export spectrum data. |
| E0066.A0003 |
SPACS |
Spectrum Digest |
[Attribute] |
27D43C |
The digital fingerprint or digest of the spectrum data. |
| E0066.A0004 |
SPACS |
YLabel |
[Attribute] |
13AB2BD |
The category or Y label of this data. Used for training supervised learning models. |
| E0066.A0005 |
SPACS |
Sequence |
[Attribute] |
1C30629 |
A compressed byte array of the spectrum data. |
| E0066.A0006 |
SPACS |
Modality |
[Attribute] |
2E160F9 |
The test/detection modality. Share the same modality enumerations with E0066.A0007. |
| E0066.A0006.V0001 |
SPACS |
XAxisMeaning |
[Value] |
34D626E |
The physiochemical meaning of X axis. E.g. for Raman, X axis means wave number. For MS, X axis means m/z or time. |
| E0066.A0007 |
SPACS |
XAxisUnit |
[Attribute] |
31AE4D2 |
X Axis Unit. e.g. cm-1 for Raman. |
| E0066.A0008 |
SPACS |
Logs |
[Attribute] |
2D6DDB4 |
Navigation property for a collection of Log objects, which tracks the historical status change of the data. |
| E0066.A0009 |
SPACS |
Spectrum Metadata |
[Attribute] |
6B802F |
Additional metadata. Can be a serialized json or xml object. |
| E0066.A0010 |
SPACS |
Spectrum Timestamp |
[Attribute] |
347B123 |
Latest revision timestamp. |
| E0071 |
SPACS |
Algorithm |
[Entity] |
1365D79 |
Represents a specific algorithm component used in the pipelines. The algorithms belong to several categories, such as baseline drift removal, averaging filter, feature scaling, feature selection, classifier, regressor, etc. |
| E0071.A0001 |
SPACS |
Algorithm Id |
[Attribute] |
146C262 |
Unique ID. Primary key. |
| E0071.A0002 |
SPACS |
Algorithm Source |
[Attribute] |
23F3797 |
The source of the algorithm. If the algorithm is self developed, source should be "private". Otherwise, specify the fully qualified module or class name, e.g. "sklearn.manifold.TSNE" |
| E0071.A0003 |
SPACS |
Algorithm Name |
[Attribute] |
3708B99 |
Name of the algorithm |
| E0071.A0004 |
SPACS |
Algorithm InputCode |
[Attribute] |
38081F1 |
Acronym or abbreviation. Used for quick search. |
| E0071.A0005 |
SPACS |
Algorithm Category |
[Attribute] |
68371 |
The category of the algorithm. Should be one of the following enumerated values. |
| E0071.A0005.V0001 |
SPACS |
Preprocessing |
[Value] |
2BE1071 |
|
| E0071.A0005.V0002 |
SPACS |
Dimension Reduction |
[Value] |
2BF0511 |
|
| E0071.A0005.V0003 |
SPACS |
Feature Selection |
[Value] |
3BC3F7C |
|
| E0071.A0005.V0004 |
SPACS |
Regression |
[Value] |
1DDB822 |
|
| E0071.A0005.V0005 |
SPACS |
Classification |
[Value] |
2839BFC |
|
| E0071.A0005.V0006 |
SPACS |
Clustering |
[Value] |
23C78CC |
|
| E0071.A0005.V0007 |
SPACS |
Visualization |
[Value] |
990B59 |
|
| E0071.A0006 |
SPACS |
Algorithm Tag |
[Attribute] |
26CBFB6 |
An additional tag for the algorithm. |
| E0071.A0007 |
SPACS |
Algorithm Reference |
[Attribute] |
292737A |
Published literature resource for the algorithm. |
| E0071.A0008 |
SPACS |
Algorithm Url |
[Attribute] |
1998A08 |
Knowledge base URL. e.g. https://en.wikipedia.org/wiki/{Name} |
| E0071.A0009 |
SPACS |
Algorithm Description |
[Attribute] |
3EFE2C8 |
A brief description for the algorithm. |
| E0071.A0010 |
SPACS |
Algorithm Metadata |
[Attribute] |
351C16B |
Metadata about the algorithm. Can be a serialized json or xml object. |
| E0071.A0011 |
SPACS |
Algorithm Implementation |
[Attribute] |
3A1382A |
The programming language or script for algorithm implementation. Should be one of the following enumerated values. |
| E0071.A0011.V0001 |
SPACS |
Python |
[Value] |
1E32406 |
|
| E0071.A0011.V0002 |
SPACS |
C/C++ |
[Value] |
204B09 |
|
| E0071.A0011.V0003 |
SPACS |
C# |
[Value] |
16DAC82 |
|
| E0071.A0011.V0004 |
SPACS |
Javascript |
[Value] |
287934C |
|
| E0071.A0011.V0005 |
SPACS |
R |
[Value] |
2577A07 |
|
| E0071.A0011.V0006 |
SPACS |
Java |
[Value] |
8D515C |
|
| E0071.A0011.V0007 |
SPACS |
Matlab |
[Value] |
249383C |
|
| E0071.A0011.V0008 |
SPACS |
Octave |
[Value] |
1CB3620 |
|
| E0071.A0012 |
SPACS |
Algorithm Code |
[Attribute] |
35B0749 |
Code snippet or pseudo code for the algorithm. |
| E0071.A0013 |
SPACS |
Algorithm Timestamp |
[Attribute] |
13A8B00 |
Latest revision timestamp. |
| E0070 |
SPACS |
Pipeline |
[Entity] |
199E3D7 |
The “Pipeline” is a set of algorithm elements organized to achieve complex data analysis tasks. A typical pipeline for spectroscopic data contains several preprocessors (e.g. filter, normalization, dimension reduction) and one regressor or classifier. |
| E0070.A0001 |
SPACS |
Pipeline Id |
[Attribute] |
4CD9E5 |
Unique ID. Primary key. |
| E0070.A0002 |
SPACS |
Pipeline Name |
[Attribute] |
38D8F0D |
Name of the pipeline |
| E0070.A0003 |
SPACS |
Pipeline InputCode |
[Attribute] |
1891537 |
Acronym or abbreviation. Used for quick search. |
| E0070.A0004 |
SPACS |
Pipeline Reference |
[Attribute] |
2E6F694 |
Literature or document describing the pipeline. |
| E0070.A0005 |
SPACS |
Pipeline Url |
[Attribute] |
1153FDD |
Knowledge base URL, which provides a preview for the pipeline. |
| E0070.A0006 |
SPACS |
Pipeline Description |
[Attribute] |
2FF1C94 |
A brief description for the pipeline. |
| E0070.A0007 |
SPACS |
Pipeline Metadata |
[Attribute] |
2149540 |
Metadata about the pipeline. Can be a serialized json or xml object. |
| E0070.A0008 |
SPACS |
Pipeline Template |
[Attribute] |
34E7FB3 |
A pipeline template that can be populated with actual data input in runtime. The current implementation uses .ipynb (IPython notebook) file as the template format. |
| E0070.A0009 |
SPACS |
Pipeline Timestamp |
[Attribute] |
423147 |
Latest revision timestamp. |
| E0068 |
SPACS |
Log |
[Entity] |
85031C |
Track the status change in the life cycle of a spectrum data. The ontology defines several phases for the spectrum data life cycle, including generate, preprocess, curate, analyze and report. |
| E0068.A0001 |
SPACS |
Log Id |
[Attribute] |
1B0385B |
Unique ID. Primary key. |
| E0068.A0002 |
SPACS |
Operator |
[Attribute] |
36A5675 |
The operator that causes the status change. Must be one of the valid users in the Account data table. |
| E0068.A0003 |
SPACS |
Operation |
[Attribute] |
1137F63 |
Should be one of the following enumerated values. |
| E0068.A0003.V0001 |
SPACS |
generate |
[Value] |
12EDDE6 |
|
| E0068.A0003.V0002 |
SPACS |
preprocess |
[Value] |
183B3C8 |
|
| E0068.A0003.V0003 |
SPACS |
curate |
[Value] |
1551A18 |
|
| E0068.A0003.V0004 |
SPACS |
analyze |
[Value] |
1180371 |
|
| E0068.A0003.V0005 |
SPACS |
report |
[Value] |
1DB9032 |
|
| E0068.A0004 |
SPACS |
Device |
[Attribute] |
4844C6 |
The instrument (e.g. MALDI-TOF-MS or Raman Spectrometer) or client computer where the operation is preformed. Should be the UID in the Device data table. |
| E0068.A0005 |
SPACS |
Location |
[Attribute] |
2CF31DD |
The institute or laboratory that performs the operation. Can also be a 3rd-party testing organization. |
| E0068.A0006 |
SPACS |
Message |
[Attribute] |
7A9EB1 |
Any messages or additional data that comes with the operation. |
| E0068.A0007 |
SPACS |
Spectrum ID |
[Attribute] |
2EE4541 |
A foreign key pointing to the related spectrum object. Spectrum and Log have one-to-many cardinality. |
| E0068.A0008 |
SPACS |
Log Timestamp |
[Attribute] |
A5B097 |
The creation timestamp of the log entry. |